Intro
I'm a medical AI researcher with extensive experience who actively contributes to the AI revolution in healthcare that is currently taking place. At Kaiko.ai we train, evaluate and apply medical foundation models. With the research team at ScreenPoint Medical, we improved the breast cancer detection performance of ScreenPoint's Transpara product. One of the areas I focused on is measuring and improving the robustness of medical AI models. I did my postdoc at Prof. Jordan Pollack's DEMO Lab at Brandeis University, and my PhD at the VUB AI Lab. Below are some examples of my work.Pathology + RNA Foundation Model
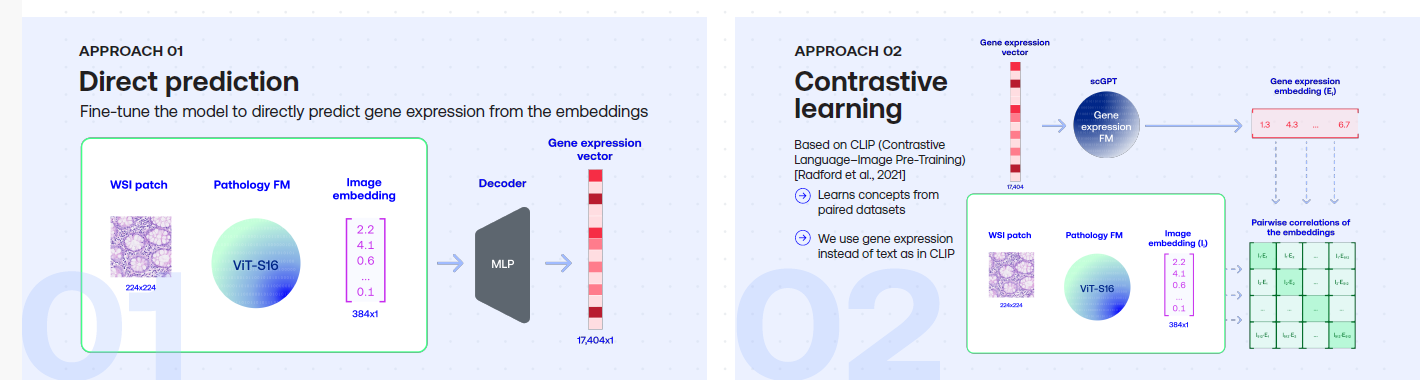
Towards Large-Scale Training of Pathology Foundation Models
To prepare for large-scale pathology FM trainings, we trained several Vision Transformer models on TCGA using DINO and DINO-v2.Paper: Towards Large-Scale Training of Pathology Foundation Models. kaiko.ai, Nanne Aben, Edwin D. de Jong, Ioannis Gatopoulos, Nicolas Känzig, Mikhail Karasikov, Axel Lagré, Roman Moser, Joost van Doorn, Fei Tang.
Foundation Model Robustness Analysis
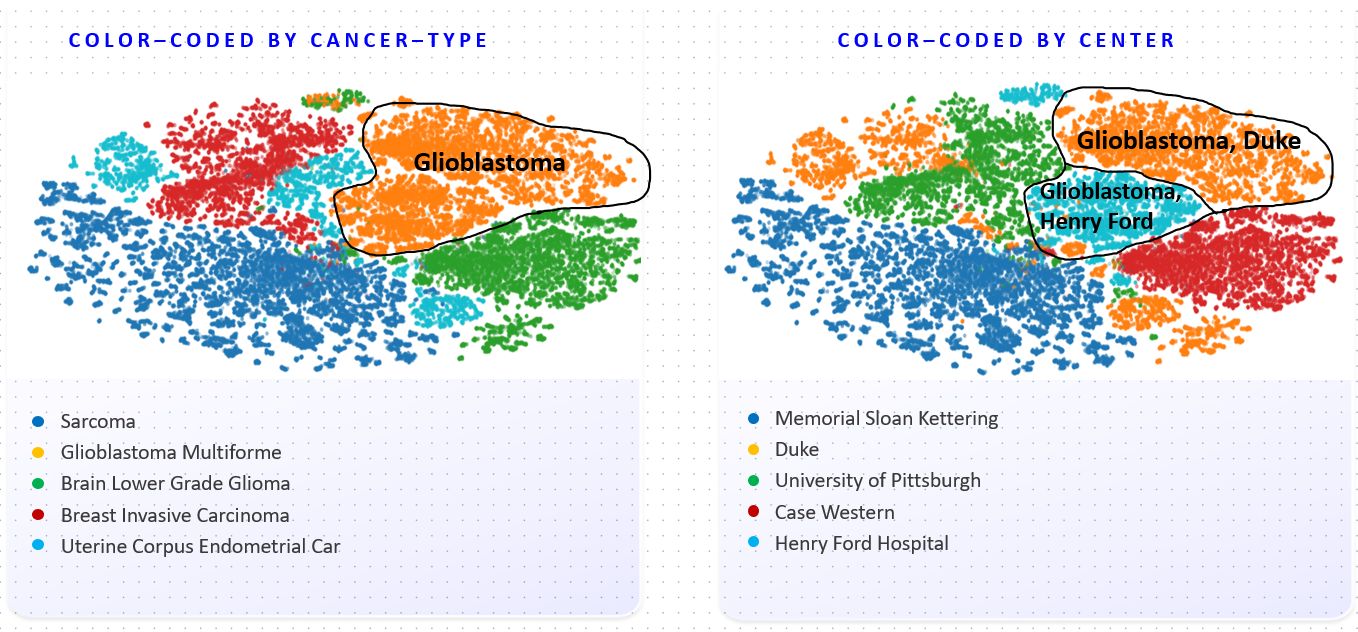
The coloring by disease shows that the FM has learned to distinguish different cancer types. The clustering on the right however shows that the overall organization of the embedding space is also strongly influenced by the medical center. This implies a risk for downstream trainings: the embeddings are not only determined by biological features such as the cancer type, but also by the center of origin, which should not play a role in downstream models and can lead to biases.
Breast Cancer Detection
With the research team at ScreenPoint Medical, we improved the breast cancer detection performance of ScreenPoint's Transpara product. The resulting model was evaluated in the large MASAI breast cancer randomized control trial in Denmark reported by Kristina Lång; see The Lancet article. This study found that our AI product reduces radiologist workload by 44%, while detecting 20% more cancer cases, and was recognized by Nature Medicine as one of the Notable advances of 2023.Incremental Sequence Learning

NIPS 2016 Workshop paper: Incremental Sequence Learning (extended version)
Incremental Sequence Learning blog post