Edwin D. de Jong
Principal Medical AI Researcher | Pathology Foundation Model Robustness | FDA Approved Radiology AI
I am a researcher who builds medical AI technology that improves healthcare.
Below I summarize my recent experience. See my selected papers below, or view my full publication list on Google Scholar  here.
here.
Experience

📄 Towards Robust Foundation Models for Digital Pathology
Invited talk on this work in the TIA seminar series:
Next, 📄 Atlas 2 was trained; a 2-billion parameter model trained on 5.5 million WSIs and currently one of the most robust pathology models globally. As the article shows, at the time of publication, Atlas 2 leads the Pareto front in performance-robustness trade-offs.
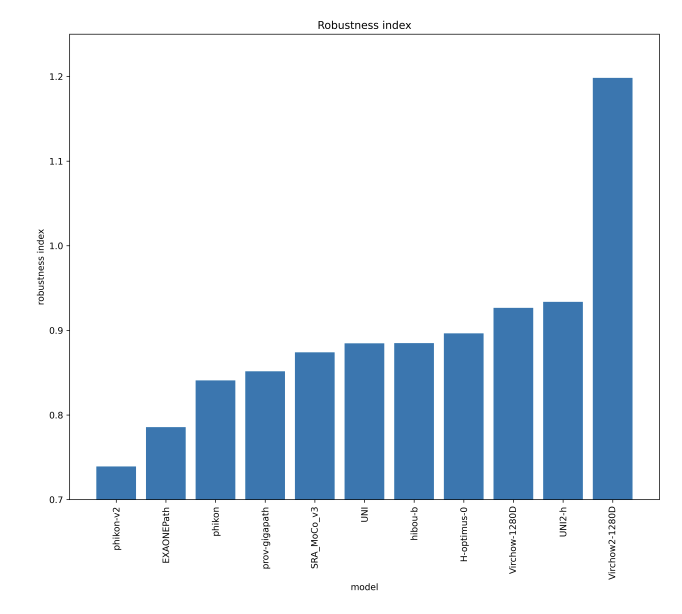
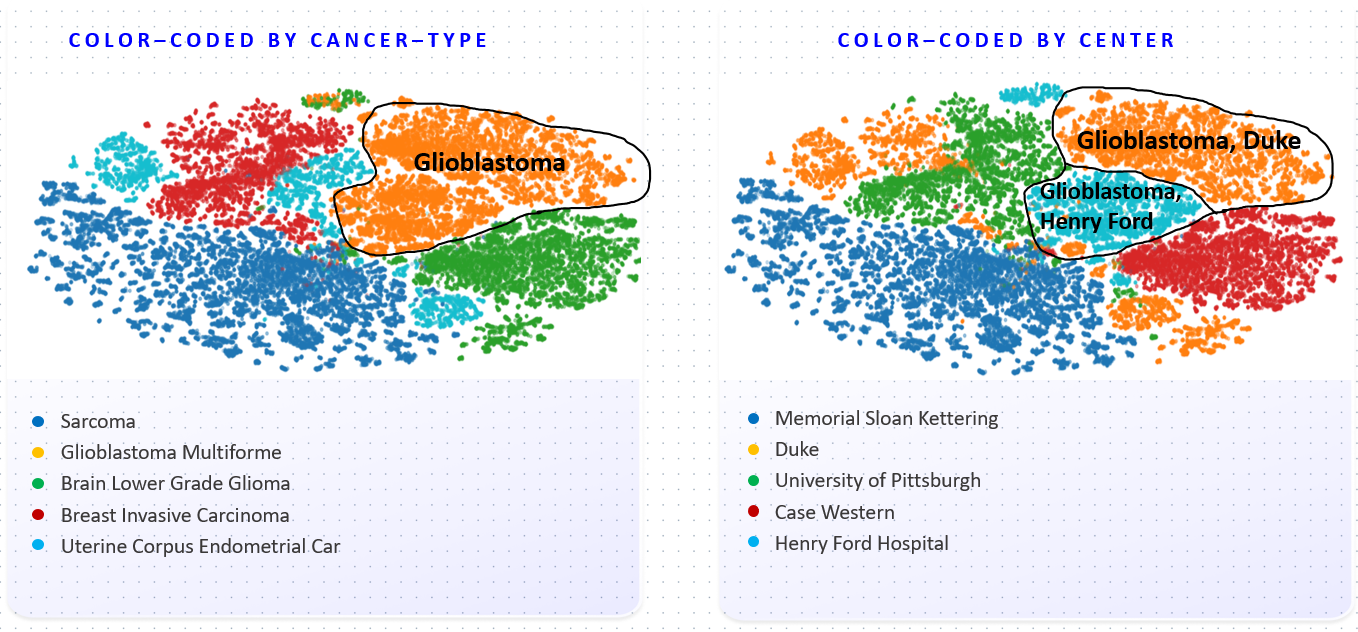
Figure explanation
Foundation Model (FM) embeddings are clustered using t-SNE and colored by disease (left) and medical center (right). Coloring by disease shows the FM has learned to distinguish cancer types. The coloring on the right however shows the embedding space also strongly represents medical centers. These should not play a role, and can lead to biases in downstream models.
📄 Current Pathology Foundation Models are unrobust to Medical Center Differences


The MASAI Randomized Controlled Trial (RCT) showed that Transpara reduces radiologist workload by 44% while at the same time increasing the cancer detection rate by 29%. It was named a Notable advance of 2023 by Nature Medicine.

Selected Papers
For a full list of research papers, see my Google Scholar page. *Asterisks denote shared first authorship.
2025
Towards Robust Foundation Models for Digital Pathology
arXiv preprint
Jonah Kömen*, Edwin D. de Jong*, Julius Hense*, Hannah Marienwald, Jonas Dippel, Philip Naumann, Eric Marcus, Lukas Ruff, Maximilian Alber, Jonas Teuwen, Frederick Klauschen, Klaus-Robert Müller
We evaluate the robustness of 20 current pathology foundation models against medical center differences. We find this lack of robustness can lead to diagnostic errors in downstream prediction models.
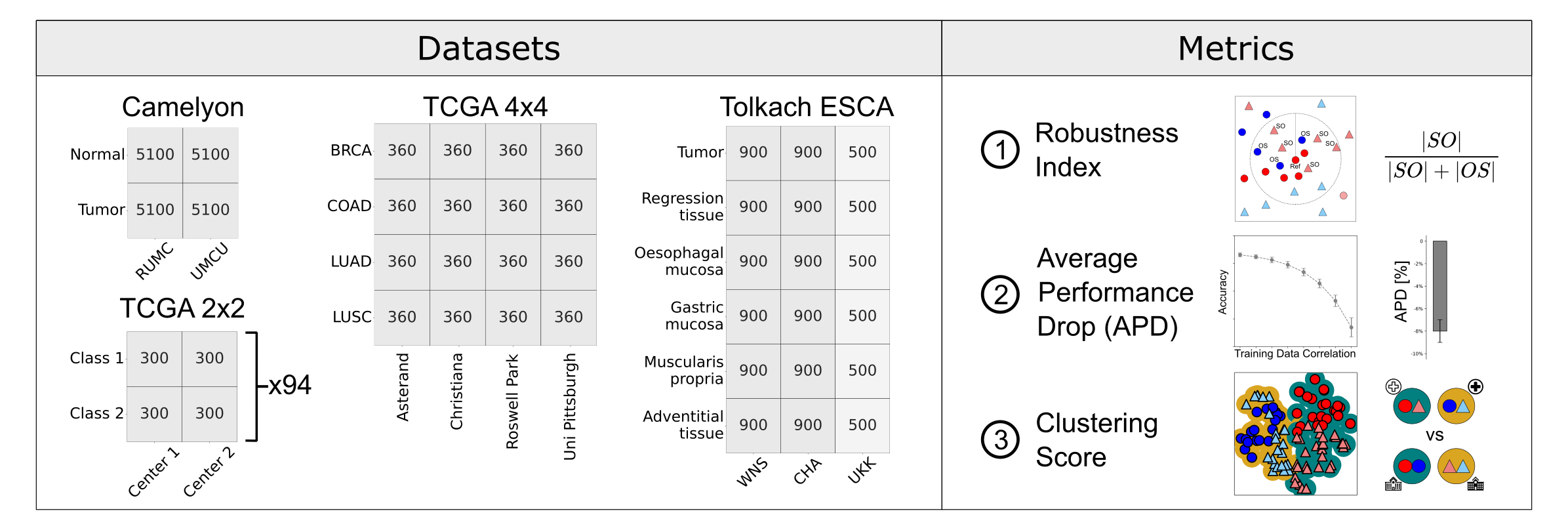
We introduce the PathoROB benchmark for evaluation of foundation model robustness, covering four datasets and 28 biological classes from 34 medical centers, and three metrics to quantify robustness.
abstract
Biomedical Foundation Models (FMs) are rapidly transforming AI-enabled healthcare research and entering clinical validation. However, their susceptibility to learning non-biological technical features -- including variations in surgical/endoscopic techniques, laboratory procedures, and scanner hardware -- poses risks for clinical deployment. We present the first systematic investigation of pathology FM robustness to non-biological features. Our work (i) introduces measures to quantify FM robustness, (ii) demonstrates the consequences of limited robustness, and (iii) proposes a framework for FM robustification to mitigate these issues. Specifically, we developed PathoROB, a robustness benchmark with three novel metrics, including the robustness index, and four datasets covering 28 biological classes from 34 medical centers. Our experiments reveal robustness deficits across all 20 evaluated FMs, and substantial robustness differences between them. We found that non-robust FM representations can cause major diagnostic downstream errors and clinical blunders that prevent safe clinical adoption. Using more robust FMs and post-hoc robustification considerably reduced (but did not yet eliminate) the risk of such errors. This work establishes that robustness evaluation is essential for validating pathology FMs before clinical adoption and demonstrates that future FM development must integrate robustness as a core design principle. PathoROB provides a blueprint for assessing robustness across biomedical domains, guiding FM improvement efforts towards more robust, representative, and clinically deployable AI systems that prioritize biological information over technical artifacts.
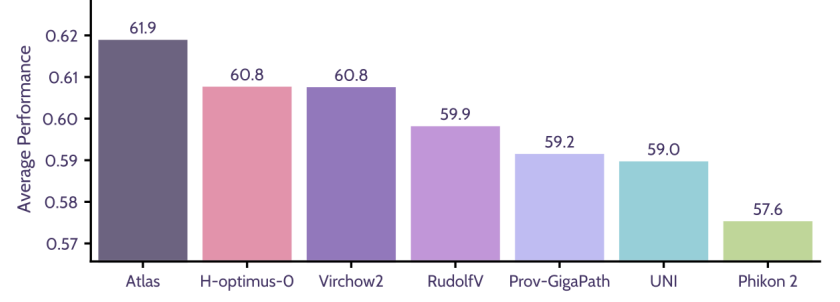
Atlas: A Novel Pathology Foundation Model by Mayo Clinic, Charité, and Aignostics
arXiv preprint
This article introduces the first Atlas foundation model.
abstract
Maximilian Alber, Stephan Tietz, Jonas Dippel, Timo Milbich, Timothée Lesort, Panos Korfiatis, Moritz Krügener, Beatriz Perez Cancer, Neelay Shah, Alexander Möllers, Philipp Seegerer, Alexandra Carpen-Amarie, Kai Standvoss, Gabriel Dernbach, Edwin de Jong, Simon Schallenberg, Andreas Kunft, Helmut Hoffer von Ankershoffen, Gavin Schaeferle, Patrick Duffy, Matt Redlon, Philipp Jurmeister, David Horst, Lukas Ruff, Klaus-Robert Müller, Frederick Klauschen, Andrew NorganRecent advances in digital pathology have demonstrated the effectiveness of foundation models across diverse applications. In this report, we present Atlas, a novel vision foundation model based on the RudolfV approach. Our model was trained on a dataset comprising 1.2 million histopathology whole slide images, collected from two medical institutions: Mayo Clinic and Charité - Universtätsmedizin Berlin. Comprehensive evaluations show that Atlas achieves state-of-the-art performance across twenty-one public benchmark datasets, even though it is neither the largest model by parameter count nor by training dataset size.
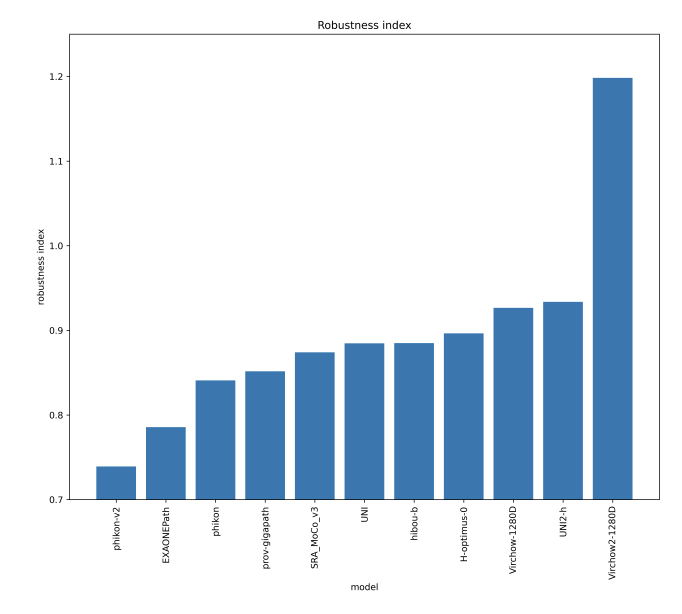
Current pathology foundation models are unrobust to medical center differences
arXiv preprint; (40+ citations within the first year)
Edwin D. de Jong, Eric Marcus, Jonas Teuwen
We measure whether pathology foundation models focus on biological features like tissue and cancer type, or on the confounding medical center signatures introduced by staining procedure and other differences. We introduce the Robustness Index: a novel metric reflecting to what degree biological features dominate confounding features.
abstract
Pathology Foundation Models (FMs) hold great promise for healthcare. Before they can be used in clinical practice, it is essential to ensure they are robust to variations between medical centers. We measure whether pathology FMs focus on biological features like tissue and cancer type, or on the well known confounding medical center signatures introduced by staining procedure and other differences. We introduce the Robustness Index. This novel robustness metric reflects to what degree biological features dominate confounding features. Ten current publicly available pathology FMs are evaluated. We find that all current pathology foundation models evaluated represent the medical center to a strong degree. Significant differences in the robustness index are observed. Only one model so far has a robustness index greater than one, meaning biological features dominate confounding features, but only slightly. A quantitative approach to measure the influence of medical center differences on FM-based prediction performance is described. We analyze the impact of unrobustness on classification performance of downstream models, and find that cancer-type classification errors are not random, but specifically attributable to same-center confounders: images of other classes from the same medical center. We visualize FM embedding spaces, and find these are more strongly organized by medical centers than by biological factors. As a consequence, the medical center of origin is predicted more accurately than the tissue source and cancer type. The robustness index introduced here is provided with the aim of advancing progress towards clinical adoption of robust and reliable pathology FMs.
2024
Towards Large-Scale Training of Pathology Foundation Models
arXiv preprint
kaiko.ai, Nanne Aben, Edwin D. de Jong, Ioannis Gatopoulos, Nicolas Känzig, Mikhail Karasikov, Axel Lagré, Roman Moser, Joost van Doorn, Fei Tang
We train a pathology foundation model on TCGA and describe our training pipeline. The eva open-source framework for pathology FM evaluation is presented.
abstract
Driven by the recent advances in deep learning methods and, in particular, by the development of modern self-supervised learning algorithms, increased interest and efforts have been devoted to build foundation models (FMs) for medical images. In this work, we present our scalable training pipeline for large pathology imaging data, and a comprehensive analysis of various hyperparameter choices and training techniques for building pathology FMs. We release and make publicly available the first batch of our pathology FMs (this https URL) trained on open-access TCGA whole slide images, a commonly used collection of pathology images. The experimental evaluation shows that our models reach state-of-the-art performance on various patch-level downstream tasks, ranging from breast cancer subtyping to colorectal nuclear segmentation. Finally, to unify the evaluation approaches used in the field and to simplify future comparisons of different FMs, we present an open-source framework (this https URL) designed for the consistent evaluation of pathology FMs across various downstream tasks.
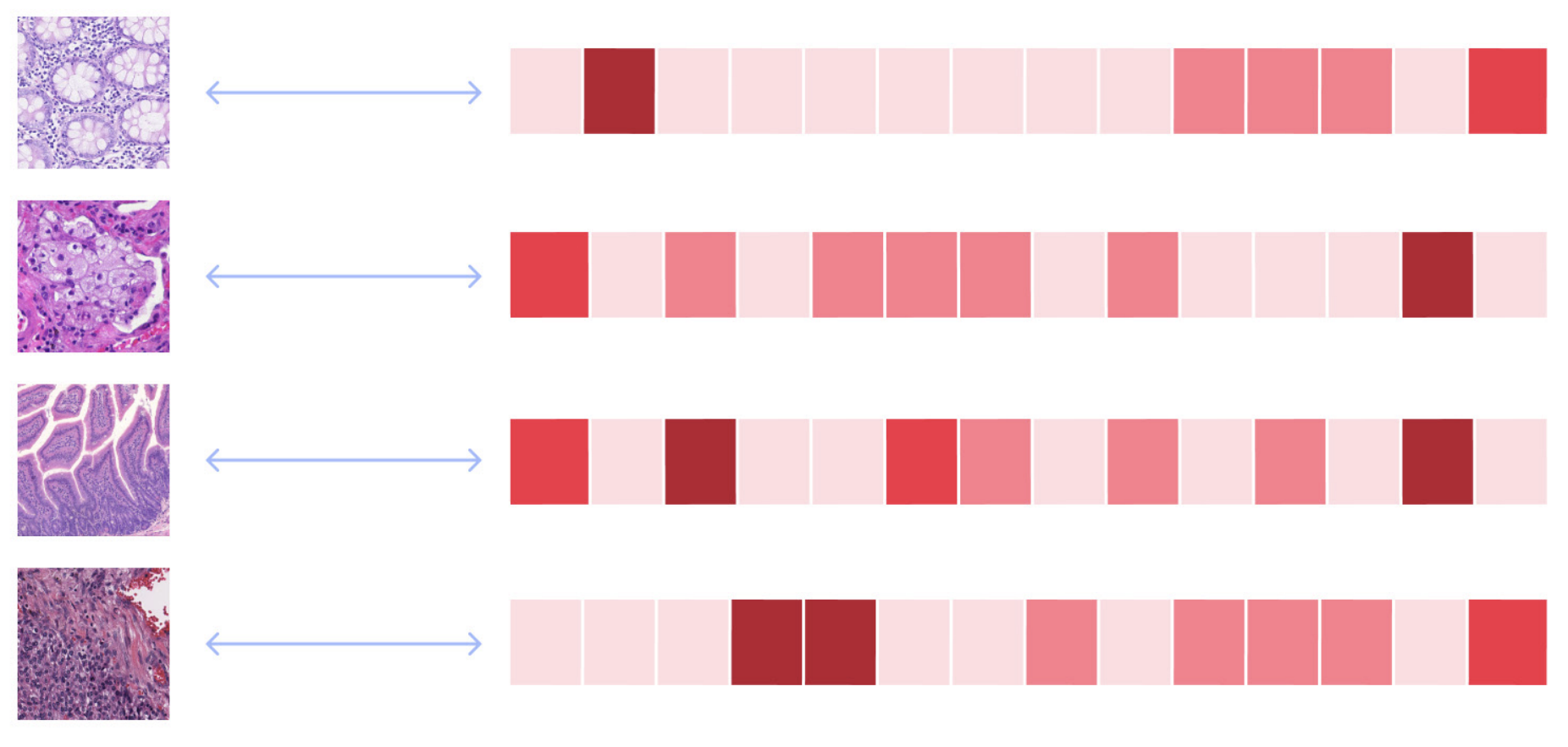
Enhancing Pathology Foundation Models with Transcriptomics.
Poster at the Genomics England Research Summit 2024.
Edwin D. de Jong, Mikhail Karasikov, Marharyta Kurban, Moritz Platscher, Marie Stettler, Fei Tang
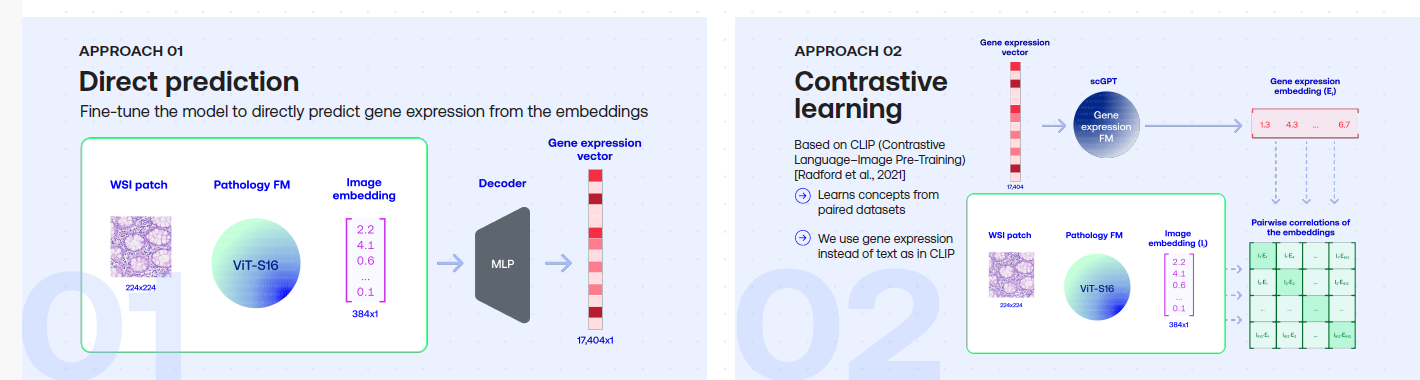
Summary
Pathology Foundation Models (FMs) such as UNI, Phikon, RudolfV and Virchow learn to represent patterns in H&E pathology images using Self-Supervised Learning. To see whether pathology FMs can be enhanced using RNA data, we compared two approaches: finetuning a pretrained pathology FM by using RNA expression levels as a high-dimensional target vector vs. using contrastive learning in a CLIP setup. We found that both approaches can improve pathology FM performance, as evaluated on a selection of downstream tasks.

Incremental sequence learning
NIPS (NeurIPS) 2016 Workshop on Continual Learning and Deep Networks
Edwin D. de Jong
Summary
This paper studies a generative Mixture Density RNN model for sequence learning. It introduces Incremental Sequence Learning: an approach to sequence learning where the length of training sequences is gradually increased over the course of training. Incremental Sequence Learning is found to speed up sequence learning by an order of magnitude, to reduce the test error, and to perform more robustly.
The paper introduces and makes available the MNIST pen stroke sequences dataset: a novel sequence learning task and data set that represents MNIST digits as sequences of pen movements that reproduce the digit.
abstract
Deep learning research over the past years has shown that by increasing the scope or difficulty of the learning problem over time, increasingly complex learning problems can be addressed. We study incremental learning in the context of sequence learning, using generative RNNs in the form of multi-layer recurrent Mixture Density Networks. While the potential of incremental or curriculum learning to enhance learning is known, indiscriminate application of the principle does not necessarily lead to improvement, and it is essential therefore to know which forms of incremental or curriculum learning have a positive effect. This research contributes to that aim by comparing three instantiations of incremental or curriculum learning.
We introduce Incremental Sequence Learning, a simple incremental approach to sequence learning. Incremental Sequence Learning starts out by using only the first few steps of each sequence as training data. Each time a performance criterion has been reached, the length of the parts of the sequences used for training is increased.
We introduce and make available a novel sequence learning task and data set: predicting and classifying MNIST pen stroke sequences. We find that Incremental Sequence Learning greatly speeds up sequence learning and reaches the best test performance level of regular sequence learning 20 times faster, reduces the test error by 74%, and in general performs more robustly; it displays lower variance and achieves sustained progress after all three comparison methods have stopped improving. The other instantiations of curriculum learning do not result in any noticeable improvement. A trained sequence prediction model is also used in transfer learning to the task of sequence classification, where it is found that transfer learning realizes improved classification performance compared to methods that learn to classify from scratch.
Projects

PathoROB benchmark: is your pathology foundation model robust?
Our work has shown that pathology foundation models (FMs) not only learn biological concepts such as organs, tissue types and types of cancer, but also represent non-biological information such as the medical center where the tissue sample was processed. Additionally, we found that such differences can lead to incorrect diagnosis predictions in downstream models.
Contact
Feel free to reach out with questions or if you’d like to discuss some of this work.
contact: jong.de.edwin.work@gmail.com, or contact me on LinkedIn  here
here
